import pandas as pd
import numpy as np
import seaborn as snsLecture 4 - Data loading and cleaning
Overview
In this lecture, we cover data loading, both from a local file and from the web. Loading data is the process of converting data (stored as a csv, HTML, etc.) into an object that Python can access and manipulate.
We also cover basic data cleaning strategies.
References
This lecture contains material from:
- Chapters 6, 7, Python for Data Analysis, 3E (Wes McKinney, 2022)
- Chapter 2, Data Science: A First Introduction with Python (Timbers et al. 2022)
- Pierre Bellec’s notes
- Chapter 5, R for Data Science (2e) (Wickham et al. 2023)
Directories
Recall from Lecture 2, we discussed your working directory - this is ‘where you are currently’ on the computer.
Python needs to know where the files live - they can be on your computer (local) or on the internet (remote).
The place where your file lives is referred to as its ‘path’. You can think of the path as directions to the file. There are two kinds of paths: relative and absolute. A relative path indicates where a file is with respect to your working directory. An absolute path indicates where the file is with respect to the computers filesystem base (or root) folder, regardless of where you are working.
Example from Data Science: A first introduction with Python (link)
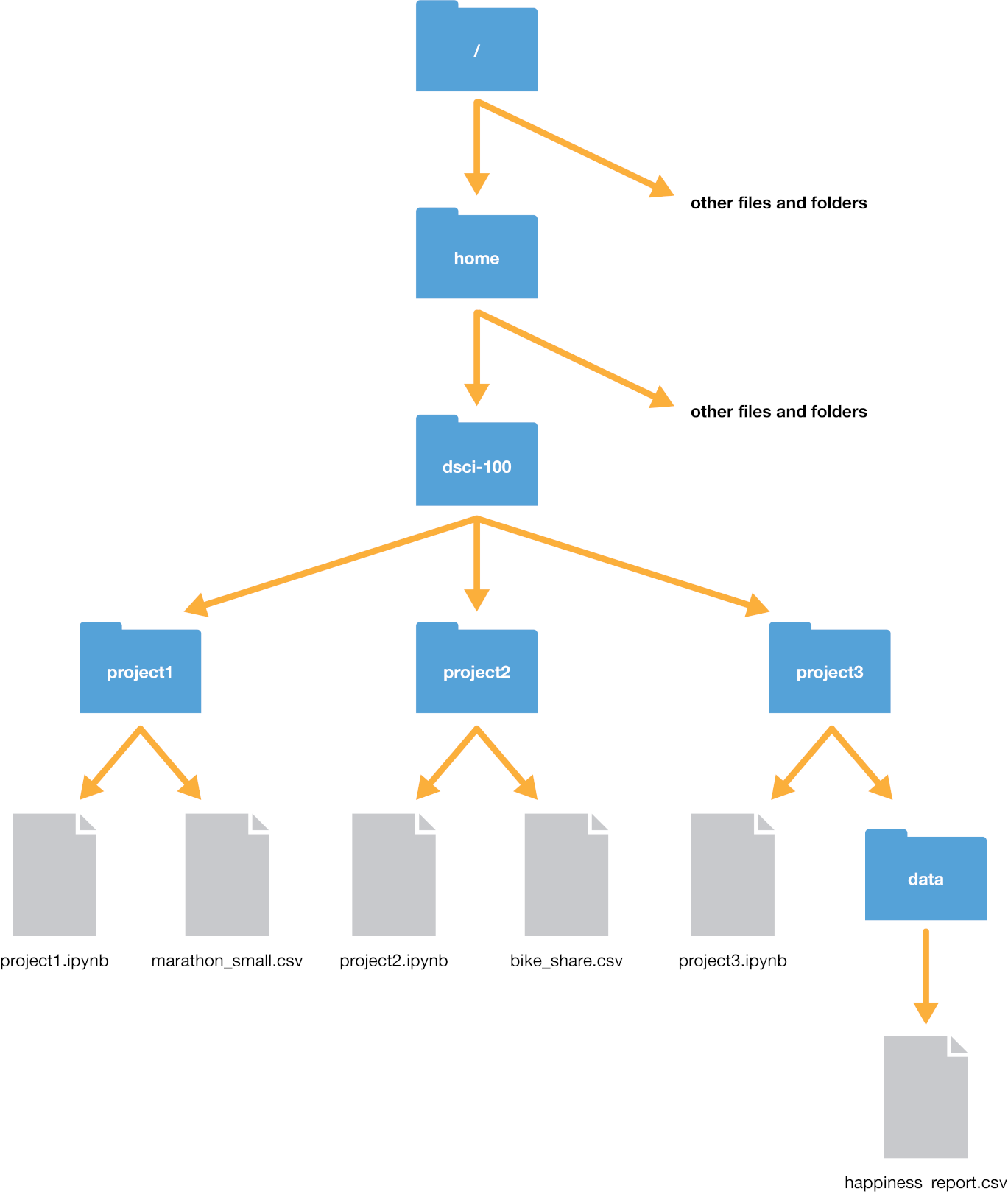
Here are some helpful shortcuts for file paths.
| Symbol | Meaning |
|---|---|
~ |
Home directory |
. |
Current working directory |
.. |
One directory up from current working directory |
../.. |
Two directories up from current working directory |
Example: suppose we are in /home/dsci-100/project3. The location of happiness_report.csv relative to our current position is: - data/happiness_report.csv (you could also use ./data/happiness_report.csv with 1 dot, instead of 2!)
Best practices
In general, you should avoid using absolute file paths in your code, as these will never work on someone else’s computer.
A good workflow for a data analysis project is:
- create a project directory (e.g.
/Users/gm845/Library/2025/project1) - in
project1, create a file structure - I like:src: this is where you put your codedata: this is where you put your datadoc: this is where you put your documents (e.g. Word or LaTex files)etc: miscellaneous files (e.g. pictures of whiteboards)
Within your project, you should use relative file paths. This makes your code portable if someone else copies your project1 folder.
In terms of naming your files, here are some good file naming practices from Jenny Bryan (link). Some key principles:
- machine readable file names (easy to narrow file lists bases on names, or extract info from file names based on splitting, NO SPACES)
- human readable file names (enough info to know what file is)
- plays well with default ordering (e.g. zero pad files 01-exercise.py, 02-exercise.py, …, 11-exercise.py)
Loading data
In Lectures 2 and 3, we looked at the exchange rate data, which we read in as a .csv (comma separated value) file using pd.read_csv('../data/rates.csv').
Comma separated values files look like this:
a,b,c,d,message
1,2,3,4,hello
5,6,7,8,world
9,10,11,12,fooex1 = pd.read_csv('../data/example_01.csv')ex1| a | b | c | d | message | |
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | hello |
| 1 | 5 | 6 | 7 | 8 | world |
| 2 | 9 | 10 | 11 | 12 | foo |
Suppose we do not have a header in the file - then, we can specify header=None:
ex2 = pd.read_csv('../data/example_02.csv', header=None)Skipping rows
!cat ../data/example_03.csvHere is a dataframe that we created:
(it is a simple example)
a,b,c,d,message
1,2,3,4,hello
5,6,7,8,world
9,10,11,12,fooNote: Above, the ! allows shell commands to be run within cells. That is, in Terminal (or another shell), we can run cat ../data/example_03.csv to display the file.
Suppose we try to load example_03.csv:
# this gives an error
# ex3 = pd.read_csv('../data/example_03.csv')We don’t want the first 2 lines; we can use the argument skiprows to skip them when loading the data:
ex3 = pd.read_csv('../data/example_03.csv', skiprows=2)ex3| a | b | c | d | message | |
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | hello |
| 1 | 5 | 6 | 7 | 8 | world |
| 2 | 9 | 10 | 11 | 12 | foo |
Using the sep argument for different separators
So far we have seen csv files. What about .txt files with tabs separating datapoints?
pd.read_csv('../data/example_04.txt', header=None)| 0 | |
|---|---|
| 0 | 1\t2\t3\t4\thello |
| 1 | 5\t6\t7\t8\tworld |
| 2 | 9\t10\t11\t12\tfoo |
ex4 = pd.read_csv('../data/example_04.txt', sep='\t', header=None)ex4| 0 | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | hello |
| 1 | 5 | 6 | 7 | 8 | world |
| 2 | 9 | 10 | 11 | 12 | foo |
Note: \t is an example of an escaped character, which always starts with a backslash (\). Escaped characters are used to represent non-printing characters (like the tab or new line \n).
Note: there are lots of potential arguments for pd.read_csv. In Lecture 2, we saw the parse_dates argument as:
df = pd.read_csv('../data/rates.csv', parse_dates=['Time'])The parse_dates argument takes a list of column names and tries to parse each of the columns as a date column.
The pd.read_csv arguments may fall into a few categories:
Indexing: Can treat one or more columns as the returned DataFrame, and whether to get column names from the file, arguments you provide, or not at all.
Type inference and data conversion: Includes the user-defined value conversions and custom list of missing value markers.
Date and time parsing: Includes a combining capability, including combining date and time information spread over multiple columns into a single column in the result.
Iterating: Support for iterating over chunks of very large files.
Unclean data issues: Includes skipping rows or a footer, comments, or other minor things like numeric data with thousands separated by commas.
For full information, you can see the documentation here.
Reading URLs
We can also read in data from a URL.
penguins = pd.read_csv("https://raw.githubusercontent.com/mwaskom/seaborn-data/master/penguins.csv")penguins.head()| species | island | bill_length_mm | bill_depth_mm | flipper_length_mm | body_mass_g | sex | |
|---|---|---|---|---|---|---|---|
| 0 | Adelie | Torgersen | 39.1 | 18.7 | 181.0 | 3750.0 | MALE |
| 1 | Adelie | Torgersen | 39.5 | 17.4 | 186.0 | 3800.0 | FEMALE |
| 2 | Adelie | Torgersen | 40.3 | 18.0 | 195.0 | 3250.0 | FEMALE |
| 3 | Adelie | Torgersen | NaN | NaN | NaN | NaN | NaN |
| 4 | Adelie | Torgersen | 36.7 | 19.3 | 193.0 | 3450.0 | FEMALE |
Reading Excel files
Gapminder is an independent education non-profit which releases public datasets (link).
We downloaded three datasets: - gdp_pcap.xlsx (Gross Domestic Product per capita) - lex.xlsx (Life expectancy) - pop.xlsx (Population)
Note: there are years past 2025 - these are projections, with details of projections in the documentation linked above.
The file name extension .xlsx refers to Microsoft Excel files, which store data very differently to csv or txt files:
PK?]Nd??xl/_rels/workbook.xml.rels<?xml version="1.0" encoding="UTF-8" standalone="yes"?>
(base) sas5ce91e6ae9a7:lec-4 gm845$ cat ../data/gdp_pcap.xlsx | head -n 2
PK?]Nd??xl/_rels/workbook.xml.rels<?xml version="1.0" encoding="UTF-8" standalone="yes"?>
<Relationships xmlns="http://schemas.openxmlformats.org/package/2006/relationships"><Relationship Id="rId1" Type="http://schemas.openxmlformats.org/officeDocument/2006/relationships/worksheet" Target="worksheets/sheet1.xml"/><Relationship Id="rId2" Type="http://schemas.openxmlformats.org/officeDocument/2006/relationships/sharedStrings" Target="sharedStrings.xml"/><Relationship Id="rId3" To read this data, we use pd.read_excel. Note: you may need to install the openpyxl package in terminal (after activating your environment):
conda activate msds597
conda install openpyxland restart the VSCode python kernel.
gdp = pd.read_excel('../data/gdp_pcap.xlsx')
pop = pd.read_excel('../data/pop.xlsx')
lex = pd.read_excel('../data/lex.xlsx')gdp.head()| country | 1800 | 1801 | 1802 | 1803 | 1804 | 1805 | 1806 | 1807 | 1808 | ... | 2091 | 2092 | 2093 | 2094 | 2095 | 2096 | 2097 | 2098 | 2099 | 2100 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | 481 | 481 | 481 | 481 | 481 | 481 | 481 | 481 | 481 | ... | 4680 | 4790 | 4910 | 5020 | 5140 | 5260 | 5380 | 5510 | 5640 | 5780 |
| 1 | Angola | 373 | 374 | 376 | 378 | 379 | 381 | 383 | 385 | 386 | ... | 24.5k | 25k | 25.6k | 26.1k | 26.6k | 27.1k | 27.7k | 28.2k | 28.8k | 29.3k |
| 2 | Albania | 469 | 471 | 472 | 473 | 475 | 476 | 477 | 479 | 480 | ... | 54.5k | 55.1k | 55.7k | 56.3k | 56.9k | 57.4k | 58k | 58.6k | 59.2k | 59.8k |
| 3 | Andorra | 1370 | 1370 | 1370 | 1380 | 1380 | 1380 | 1390 | 1390 | 1390 | ... | 79.9k | 80.2k | 80.4k | 80.7k | 81k | 81.3k | 81.5k | 81.8k | 82k | 82.3k |
| 4 | UAE | 1140 | 1150 | 1150 | 1150 | 1160 | 1160 | 1170 | 1170 | 1180 | ... | 92.6k | 92.6k | 92.6k | 92.7k | 92.7k | 92.7k | 92.8k | 92.8k | 92.8k | 92.9k |
5 rows × 302 columns
pop.head()| country | 1800 | 1801 | 1802 | 1803 | 1804 | 1805 | 1806 | 1807 | 1808 | ... | 2091 | 2092 | 2093 | 2094 | 2095 | 2096 | 2097 | 2098 | 2099 | 2100 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | 3.28M | 3.28M | 3.28M | 3.28M | 3.28M | 3.28M | 3.28M | 3.28M | 3.28M | ... | 124M | 125M | 126M | 126M | 127M | 128M | 128M | 129M | 130M | 130M |
| 1 | Angola | 1.57M | 1.57M | 1.57M | 1.57M | 1.57M | 1.57M | 1.57M | 1.57M | 1.57M | ... | 139M | 140M | 142M | 143M | 144M | 145M | 147M | 148M | 149M | 150M |
| 2 | Albania | 400k | 402k | 404k | 405k | 407k | 409k | 411k | 413k | 414k | ... | 1.34M | 1.32M | 1.3M | 1.29M | 1.27M | 1.25M | 1.23M | 1.22M | 1.2M | 1.18M |
| 3 | Andorra | 2650 | 2650 | 2650 | 2650 | 2650 | 2650 | 2650 | 2650 | 2650 | ... | 52.8k | 52.1k | 51.5k | 50.8k | 50.2k | 49.6k | 49k | 48.4k | 47.8k | 47.2k |
| 4 | UAE | 40.2k | 40.2k | 40.2k | 40.2k | 40.2k | 40.2k | 40.2k | 40.2k | 40.2k | ... | 24.1M | 24.3M | 24.5M | 24.7M | 25M | 25.2M | 25.4M | 25.7M | 25.9M | 26.1M |
5 rows × 302 columns
lex.head()| country | 1800 | 1801 | 1802 | 1803 | 1804 | 1805 | 1806 | 1807 | 1808 | ... | 2091 | 2092 | 2093 | 2094 | 2095 | 2096 | 2097 | 2098 | 2099 | 2100 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | 28.2 | 28.2 | 28.2 | 28.2 | 28.2 | 28.2 | 28.1 | 28.1 | 28.1 | ... | 75.9 | 76.1 | 76.2 | 76.4 | 76.5 | 76.7 | 76.8 | 77.0 | 77.1 | 77.3 |
| 1 | Angola | 27.0 | 27.0 | 27.0 | 27.0 | 27.0 | 27.0 | 27.0 | 27.0 | 27.0 | ... | 75.8 | 75.9 | 76.0 | 76.1 | 76.3 | 76.4 | 76.5 | 76.7 | 76.8 | 76.9 |
| 2 | Albania | 35.4 | 35.4 | 35.4 | 35.4 | 35.4 | 35.4 | 35.4 | 35.4 | 35.4 | ... | 88.9 | 89.0 | 89.2 | 89.3 | 89.4 | 89.5 | 89.6 | 89.7 | 89.8 | 90.0 |
| 3 | Andorra | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | ... | 91.0 | 91.1 | 91.3 | 91.4 | 91.5 | 91.6 | 91.7 | 91.8 | 91.9 | 92.0 |
| 4 | UAE | 30.7 | 30.7 | 30.7 | 30.7 | 30.7 | 30.7 | 30.7 | 30.7 | 30.7 | ... | 83.7 | 83.8 | 83.9 | 84.0 | 84.1 | 84.2 | 84.3 | 84.5 | 84.6 | 84.7 |
5 rows × 302 columns
Let’s practice some of our data skills from previous lectures.
- Turn each dataset into tidy data
gdp = gdp.melt(id_vars='country',
var_name='year',
value_name='gdp')gdp.head()| country | year | gdp | |
|---|---|---|---|
| 0 | Afghanistan | 1800 | 481 |
| 1 | Angola | 1800 | 373 |
| 2 | Albania | 1800 | 469 |
| 3 | Andorra | 1800 | 1370 |
| 4 | UAE | 1800 | 1140 |
pop = pop.melt(id_vars = 'country',
var_name = 'year',
value_name = 'pop')
lex = lex.melt(id_vars = 'country',
var_name = 'year',
value_name = 'lex')- Merge the datasets together
gapminder = gdp.merge(pop, on=['country', 'year']).merge(lex, on=['country', 'year'])gapminder.head()| country | year | gdp | pop | lex | |
|---|---|---|---|---|---|
| 0 | Afghanistan | 1800 | 481 | 3.28M | 28.2 |
| 1 | Angola | 1800 | 373 | 1.57M | 27.0 |
| 2 | Albania | 1800 | 469 | 400k | 35.4 |
| 3 | Andorra | 1800 | 1370 | 2650 | NaN |
| 4 | UAE | 1800 | 1140 | 40.2k | 30.7 |
- Keep only years 1950-2019
keep = (gapminder['year'] >= 1950) & (gapminder['year'] <= 2019)# an alternative
keep = gapminder['year'].between(1950, 2019)gapminder = gapminder[keep]gapminder = gapminder.reset_index().drop(columns='index')- Are there any NaNs?
gapminder.isna().sum()country 0
year 0
gdp 0
pop 0
lex 0
dtype: int64String manipulation
Illustration: Gapminder data
We can still see some issues - numbers are in 14M and 14k formats and are read as strings, not integers or floats.
We can use Python string methods to clean this data.
First, let’s look at some of the data:
test = gapminder.loc[0, 'pop']test'7.78M'isinstance(test, str)TrueNumbers in the millions are stored as strings.
test2 = gapminder.loc[3,'pop']test26000type(test2)intWe can iterate over strings:
for ch in test:
print(ch)7
.
7
8
MFor each string element, we can see if it’s a letter using .isalpha().
for ch in test:
if ch.isalpha():
print(ch)MFinally, let’s write our function that returns all the letters we may see in the pd.Series.
def get_number_formats(x):
num_forms = []
for val in x:
if isinstance(val, str):
for ch in val:
if ch.isalpha():
num_forms.append(ch)
return set(num_forms)num_forms = get_number_formats(gapminder['pop'])num_forms{'B', 'M', 'k'}Now, convert the strings to floats:
def convert_to_float(x):
if isinstance(x, str):
if 'k' in x:
x_new = float(x.replace('k', '')) * 1000
if 'M' in x:
x_new = float(x.replace('M', '')) * 1000000
if 'B' in x:
x_new = float(x.replace('B', '')) * 1000000000
return x_new
else:
return xgapminder[['gdp', 'pop']] = gapminder[['gdp', 'pop']].map(convert_to_float)gapminder['pop'] = gapminder['pop'].astype('int64')
gapminder['year'] = gapminder['year'].astype('int64')year_plot = 2007
g = sns.relplot(gapminder[gapminder['year'] == year_plot],
x='gdp',
y='lex',
size='pop',
height = 4,
aspect = 2)
g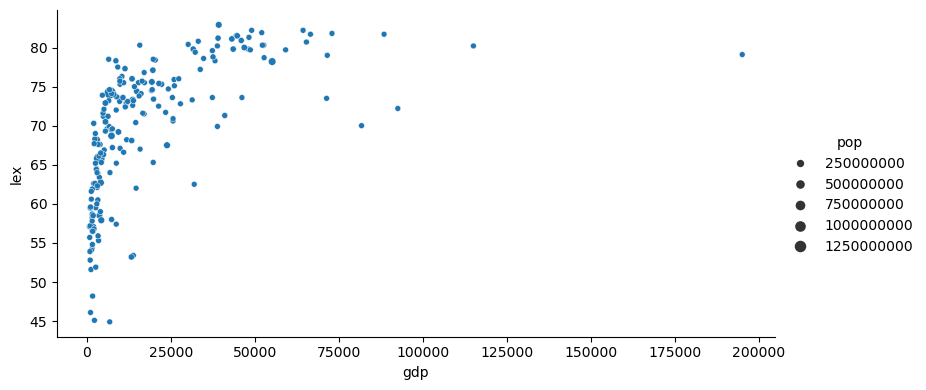
Let’s plot in log scale for the x-axis:
g = sns.relplot(gapminder[gapminder['year'] == year_plot],
x='gdp',
y='lex',
size='pop',
height = 4,
aspect = 2)
g.set(xscale='log')
g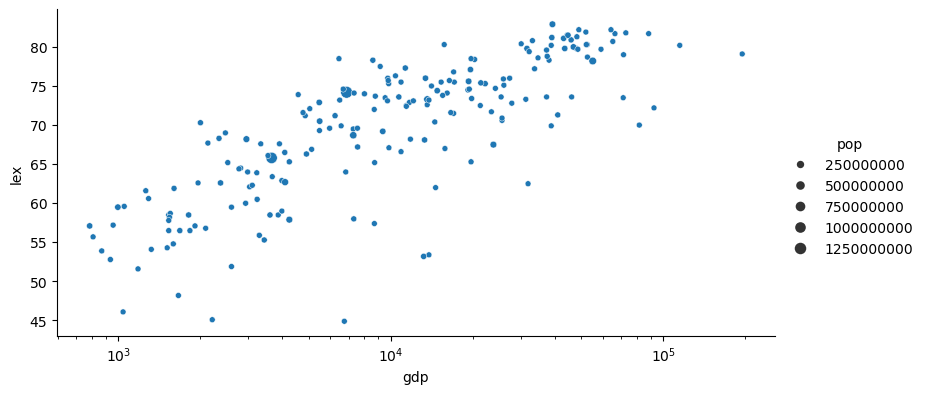
Above, we used the following Python built-in string methods:
.isalpha.replace
Another helpful method is .split(), which breaks strings into a list of substrings using the passed delimiter:
my_string = '2,3,4,5'
my_string.split(',')['2', '3', '4', '5']String functions in pandas
We can apply string methods to pandas dataframes using pd.DataFrame.str.<method>.
Here is a list of some more helpful functions.
subset = gapminder['country'].str.contains('U')gapminder['country'][subset].unique()array(['UAE', 'UK', 'Uganda', 'Ukraine', 'Uruguay', 'USA', 'Uzbekistan'],
dtype=object)Standardizing Country Names
Let’s go back to our gapminder data! We are going to add continent data.
Install these packages in your msds597 environment:
Terminal
pip install pycountry
pip install pycountry_convertimport pycountry
import pycountry_convert as pcdf_country = gapminder[['country']].drop_duplicates()df_country['country_code'] = pd.NAdf_country.dtypescountry object
country_code object
dtype: objectexceptions = []
for c in df_country['country']:
try:
df_country.loc[df_country['country']==c, 'country_code'] = pc.country_name_to_country_alpha2(c)
except:
exceptions.append(c)exceptions['UAE',
"Cote d'Ivoire",
'Congo, Dem. Rep.',
'Congo, Rep.',
'Micronesia, Fed. Sts.',
'UK',
'Hong Kong, China',
'Lao',
'St. Vincent and the Grenadines']Let’s now do the painstaking work of finding exceptions:
uae = pycountry.countries.search_fuzzy('emirates')uae[Country(alpha_2='AE', alpha_3='ARE', flag='🇦🇪', name='United Arab Emirates', numeric='784')]df_country['country'].str.contains('UAE')0 False
1 False
2 False
3 False
4 True
...
190 False
191 False
192 False
193 False
194 False
Name: country, Length: 195, dtype: booldf_country.loc[df_country['country'].str.contains('UAE'), 'country_code'] = uae[0].alpha_2ivoire = pycountry.countries.search_fuzzy('ivoire')df_country.loc[df_country['country'].str.contains('Ivoire'), 'country_code'] = ivoire[0].alpha_2congo = pycountry.countries.search_fuzzy('congo')congo[Country(alpha_2='CG', alpha_3='COG', flag='🇨🇬', name='Congo', numeric='178', official_name='Republic of the Congo'),
Country(alpha_2='CD', alpha_3='COD', flag='🇨🇩', name='Congo, The Democratic Republic of the', numeric='180')]df_country.loc[df_country['country'].str.contains('Congo, Dem.'), 'country_code'] = congo[1].alpha_2df_country.loc[df_country['country'].str.contains('Congo, Rep.'), 'country_code'] = congo[0].alpha_2mic = pycountry.countries.search_fuzzy('micronesia')df_country.loc[df_country['country'].str.contains('Micronesia, Fed. Sts.'), 'country_code'] = mic[0].alpha_2gb = pycountry.countries.search_fuzzy('great britain')df_country.loc[df_country['country'].str.contains('UK'), 'country_code'] = gb[0].alpha_2hk = pycountry.countries.search_fuzzy('hong kong')df_country.loc[df_country['country'].str.contains('Hong Kong'), 'country_code'] = hk[0].alpha_2laos = pycountry.countries.search_fuzzy('Laos')df_country.loc[df_country['country'].str.contains('Lao'), 'country_code'] = laos[0].alpha_2vin = pycountry.countries.search_fuzzy('Vincent')df_country.loc[df_country['country'].str.contains('Vincent'), 'country_code'] = vin[0].alpha_2Have we found them all?
any(df_country['country_code'].isna())Falsedf_country['continent'] = pd.NAexceptions = []
for c in df_country['country_code']:
try:
cc = pc.country_alpha2_to_continent_code(c)
df_country.loc[df_country['country_code']==c, 'continent'] = pc.convert_continent_code_to_continent_name(cc)
except:
exceptions.append(c)exceptions['TL']df_country.loc[df_country['country_code']=='TL', 'continent'] = 'Asia'any(df_country['continent'].isna())FalseMerging continent data
gapminder = gapminder.merge(df_country, on='country')gapminder.head()| country | year | gdp | pop | lex | country_code | continent | |
|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | 1950 | 1450.0 | 7780000 | 42.7 | AF | Asia |
| 1 | Angola | 1950 | 2230.0 | 4550000 | 45.6 | AO | Africa |
| 2 | Albania | 1950 | 1980.0 | 1250000 | 52.2 | AL | Europe |
| 3 | Andorra | 1950 | 8350.0 | 6000 | 74.6 | AD | Europe |
| 4 | UAE | 1950 | 1710.0 | 74500 | 58.4 | AE | Asia |
g = sns.relplot(gapminder[gapminder['year'] == year_plot],
x='gdp',
y='lex',
size='pop',
hue='continent',
height = 4.5,
aspect = 1.5)
g.set(xscale='log')
g.set_axis_labels("Gross Domestic Product per Capita", "Life Expectancy")
g.set(title=f'Gapminder Data (Year = {year_plot})')
g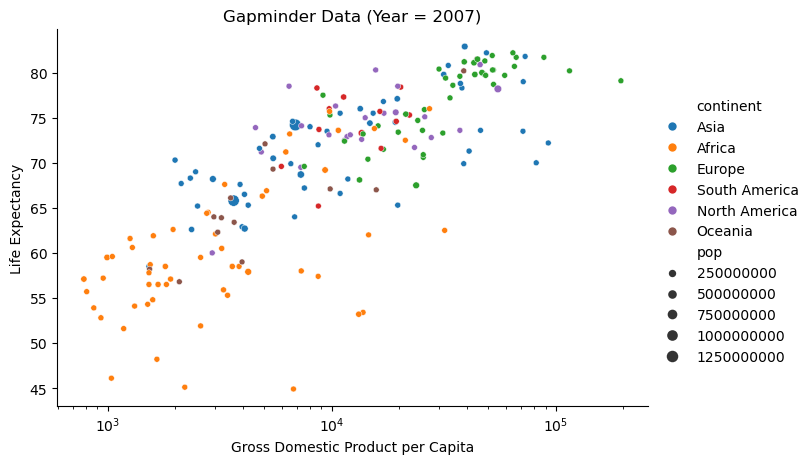
Illustration: World Health Organization Tuberculosis Data
We now consider another data cleaning exercise, adapted from R For Data Science (2e).
The data is who_data.csv, collected by the World Health Organisation (link). It contains information about tuberculosis diagnoses. The data contains:
- country
- year
- 56 columns like
new_sp_m_014,new_ep_m_4554, andnew_rel_m_3544.
Each of the 56 columns follow a pattern.
The first three letters of each column denote whether the column contains new or old cases of TB. In this dataset, each column contains new cases.
The next two letters describe the type of TB:
relstands for cases of relapseepstands for cases of extrapulmonary TBsnstands for cases of pulmonary TB that could not be diagnosed by a pulmonary smear (smear negative)spstands for cases of pulmonary TB that could be diagnosed by a pulmonary smear (smear positive)
The sixth letter gives the sex of TB patients. The dataset groups cases by males (m) and females (f).
The remaining numbers gives the age group. The dataset groups cases into seven age groups:
- 014 = 0 – 14 years old
- 1524 = 15 – 24 years old
- 2534 = 25 – 34 years old
- 3544 = 35 – 44 years old
- 4554 = 45 – 54 years old
- 5564 = 55 – 64 years old
- 65 = 65 or older
who = pd.read_csv('../data/who_data.csv')
who.head()| country | iso2 | iso3 | year | new_sp_m014 | new_sp_m1524 | new_sp_m2534 | new_sp_m3544 | new_sp_m4554 | new_sp_m5564 | ... | newrel_m4554 | newrel_m5564 | newrel_m65 | newrel_f014 | newrel_f1524 | newrel_f2534 | newrel_f3544 | newrel_f4554 | newrel_f5564 | newrel_f65 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | Afghanistan | AF | AFG | 1980 | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 1 | Afghanistan | AF | AFG | 1981 | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 2 | Afghanistan | AF | AFG | 1982 | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 3 | Afghanistan | AF | AFG | 1983 | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
| 4 | Afghanistan | AF | AFG | 1984 | NaN | NaN | NaN | NaN | NaN | NaN | ... | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN | NaN |
5 rows × 60 columns
Let’s first put the data in tidy form:
who_melt = who.melt(id_vars=['country', 'iso2', 'iso3', 'year'],
var_name='key',
value_name='cases')who_melt.head()| country | iso2 | iso3 | year | key | cases | |
|---|---|---|---|---|---|---|
| 0 | Afghanistan | AF | AFG | 1980 | new_sp_m014 | NaN |
| 1 | Afghanistan | AF | AFG | 1981 | new_sp_m014 | NaN |
| 2 | Afghanistan | AF | AFG | 1982 | new_sp_m014 | NaN |
| 3 | Afghanistan | AF | AFG | 1983 | new_sp_m014 | NaN |
| 4 | Afghanistan | AF | AFG | 1984 | new_sp_m014 | NaN |
Let’s drop NaNs.
who_melt = who_melt.dropna()
who_melt.head()| country | iso2 | iso3 | year | key | cases | |
|---|---|---|---|---|---|---|
| 17 | Afghanistan | AF | AFG | 1997 | new_sp_m014 | 0.0 |
| 18 | Afghanistan | AF | AFG | 1998 | new_sp_m014 | 30.0 |
| 19 | Afghanistan | AF | AFG | 1999 | new_sp_m014 | 8.0 |
| 20 | Afghanistan | AF | AFG | 2000 | new_sp_m014 | 52.0 |
| 21 | Afghanistan | AF | AFG | 2001 | new_sp_m014 | 129.0 |
who_melt['key'].unique()array(['new_sp_m014', 'new_sp_m1524', 'new_sp_m2534', 'new_sp_m3544',
'new_sp_m4554', 'new_sp_m5564', 'new_sp_m65', 'new_sp_f014',
'new_sp_f1524', 'new_sp_f2534', 'new_sp_f3544', 'new_sp_f4554',
'new_sp_f5564', 'new_sp_f65', 'new_sn_m014', 'new_sn_m1524',
'new_sn_m2534', 'new_sn_m3544', 'new_sn_m4554', 'new_sn_m5564',
'new_sn_m65', 'new_sn_f014', 'new_sn_f1524', 'new_sn_f2534',
'new_sn_f3544', 'new_sn_f4554', 'new_sn_f5564', 'new_sn_f65',
'new_ep_m014', 'new_ep_m1524', 'new_ep_m2534', 'new_ep_m3544',
'new_ep_m4554', 'new_ep_m5564', 'new_ep_m65', 'new_ep_f014',
'new_ep_f1524', 'new_ep_f2534', 'new_ep_f3544', 'new_ep_f4554',
'new_ep_f5564', 'new_ep_f65', 'newrel_m014', 'newrel_m1524',
'newrel_m2534', 'newrel_m3544', 'newrel_m4554', 'newrel_m5564',
'newrel_m65', 'newrel_f014', 'newrel_f1524', 'newrel_f2534',
'newrel_f3544', 'newrel_f4554', 'newrel_f5564', 'newrel_f65'],
dtype=object)Looks like most keys have pattern new_type_sexage, except newrel. Let’s change that.
who_melt['key'] = who_melt['key'].str.replace('newrel', 'new_rel')who_melt['key'].unique()array(['new_sp_m014', 'new_sp_m1524', 'new_sp_m2534', 'new_sp_m3544',
'new_sp_m4554', 'new_sp_m5564', 'new_sp_m65', 'new_sp_f014',
'new_sp_f1524', 'new_sp_f2534', 'new_sp_f3544', 'new_sp_f4554',
'new_sp_f5564', 'new_sp_f65', 'new_sn_m014', 'new_sn_m1524',
'new_sn_m2534', 'new_sn_m3544', 'new_sn_m4554', 'new_sn_m5564',
'new_sn_m65', 'new_sn_f014', 'new_sn_f1524', 'new_sn_f2534',
'new_sn_f3544', 'new_sn_f4554', 'new_sn_f5564', 'new_sn_f65',
'new_ep_m014', 'new_ep_m1524', 'new_ep_m2534', 'new_ep_m3544',
'new_ep_m4554', 'new_ep_m5564', 'new_ep_m65', 'new_ep_f014',
'new_ep_f1524', 'new_ep_f2534', 'new_ep_f3544', 'new_ep_f4554',
'new_ep_f5564', 'new_ep_f65', 'new_rel_m014', 'new_rel_m1524',
'new_rel_m2534', 'new_rel_m3544', 'new_rel_m4554', 'new_rel_m5564',
'new_rel_m65', 'new_rel_f014', 'new_rel_f1524', 'new_rel_f2534',
'new_rel_f3544', 'new_rel_f4554', 'new_rel_f5564', 'new_rel_f65'],
dtype=object)How to split up these keys?
who_melt['key'].str.split('_')17 [new, sp, m014]
18 [new, sp, m014]
19 [new, sp, m014]
20 [new, sp, m014]
21 [new, sp, m014]
...
405269 [new, rel, f65]
405303 [new, rel, f65]
405371 [new, rel, f65]
405405 [new, rel, f65]
405439 [new, rel, f65]
Name: key, Length: 75752, dtype: objectwho_split = who_melt['key'].str.split('_', expand=True)
who_split| 0 | 1 | 2 | |
|---|---|---|---|
| 17 | new | sp | m014 |
| 18 | new | sp | m014 |
| 19 | new | sp | m014 |
| 20 | new | sp | m014 |
| 21 | new | sp | m014 |
| ... | ... | ... | ... |
| 405269 | new | rel | f65 |
| 405303 | new | rel | f65 |
| 405371 | new | rel | f65 |
| 405405 | new | rel | f65 |
| 405439 | new | rel | f65 |
75752 rows × 3 columns
Change column names:
who_split.columns = ['new', 'type', 'sexage']Split sexage into sex and age columns:
who_split['sexage'].str.slice(start=0, stop=1)17 m
18 m
19 m
20 m
21 m
..
405269 f
405303 f
405371 f
405405 f
405439 f
Name: sexage, Length: 75752, dtype: objectwho_split['sex'] = who_split['sexage'].str.slice(start=0, stop=1)who_split['age'] = who_split['sexage'].str.slice(start=1)who_split.head()| new | type | sexage | sex | age | |
|---|---|---|---|---|---|
| 17 | new | sp | m014 | m | 014 |
| 18 | new | sp | m014 | m | 014 |
| 19 | new | sp | m014 | m | 014 |
| 20 | new | sp | m014 | m | 014 |
| 21 | new | sp | m014 | m | 014 |
| ... | ... | ... | ... | ... | ... |
| 405269 | new | rel | f65 | f | 65 |
| 405303 | new | rel | f65 | f | 65 |
| 405371 | new | rel | f65 | f | 65 |
| 405405 | new | rel | f65 | f | 65 |
| 405439 | new | rel | f65 | f | 65 |
75752 rows × 5 columns
In this dataset, all cases are new:
who_split['new'].unique()array(['new'], dtype=object)who_split = who_split.drop(columns=['new', 'sexage'])who_split['age'] = who_split['age'].astype('category')who_split['age'] = who_split['age'].cat.as_ordered()who_split['age']17 014
18 014
19 014
20 014
21 014
...
405269 65
405303 65
405371 65
405405 65
405439 65
Name: age, Length: 75752, dtype: category
Categories (7, object): ['014' < '1524' < '2534' < '3544' < '4554' < '5564' < '65']who_split.head()| type | sex | age | |
|---|---|---|---|
| 17 | sp | m | 014 |
| 18 | sp | m | 014 |
| 19 | sp | m | 014 |
| 20 | sp | m | 014 |
| 21 | sp | m | 014 |
who_tidy = who_melt.merge(who_split, left_index=True, right_index=True)
who_tidy.head()| country | iso2 | iso3 | year | key | cases | type | sex | age | |
|---|---|---|---|---|---|---|---|---|---|
| 17 | Afghanistan | AF | AFG | 1997 | new_sp_m014 | 0.0 | sp | m | 014 |
| 18 | Afghanistan | AF | AFG | 1998 | new_sp_m014 | 30.0 | sp | m | 014 |
| 19 | Afghanistan | AF | AFG | 1999 | new_sp_m014 | 8.0 | sp | m | 014 |
| 20 | Afghanistan | AF | AFG | 2000 | new_sp_m014 | 52.0 | sp | m | 014 |
| 21 | Afghanistan | AF | AFG | 2001 | new_sp_m014 | 129.0 | sp | m | 014 |
who_tidy = who_tidy.drop(columns=['key'])who_tidy.head()| country | iso2 | iso3 | year | cases | type | sex | age | |
|---|---|---|---|---|---|---|---|---|
| 17 | Afghanistan | AF | AFG | 1997 | 0.0 | sp | m | 014 |
| 18 | Afghanistan | AF | AFG | 1998 | 30.0 | sp | m | 014 |
| 19 | Afghanistan | AF | AFG | 1999 | 8.0 | sp | m | 014 |
| 20 | Afghanistan | AF | AFG | 2000 | 52.0 | sp | m | 014 |
| 21 | Afghanistan | AF | AFG | 2001 | 129.0 | sp | m | 014 |
sns.relplot(who_tidy[who_tidy.iso2 == 'AF'],
x='year',
y='cases',
hue='type',
style='sex',
col='age',
col_wrap=3)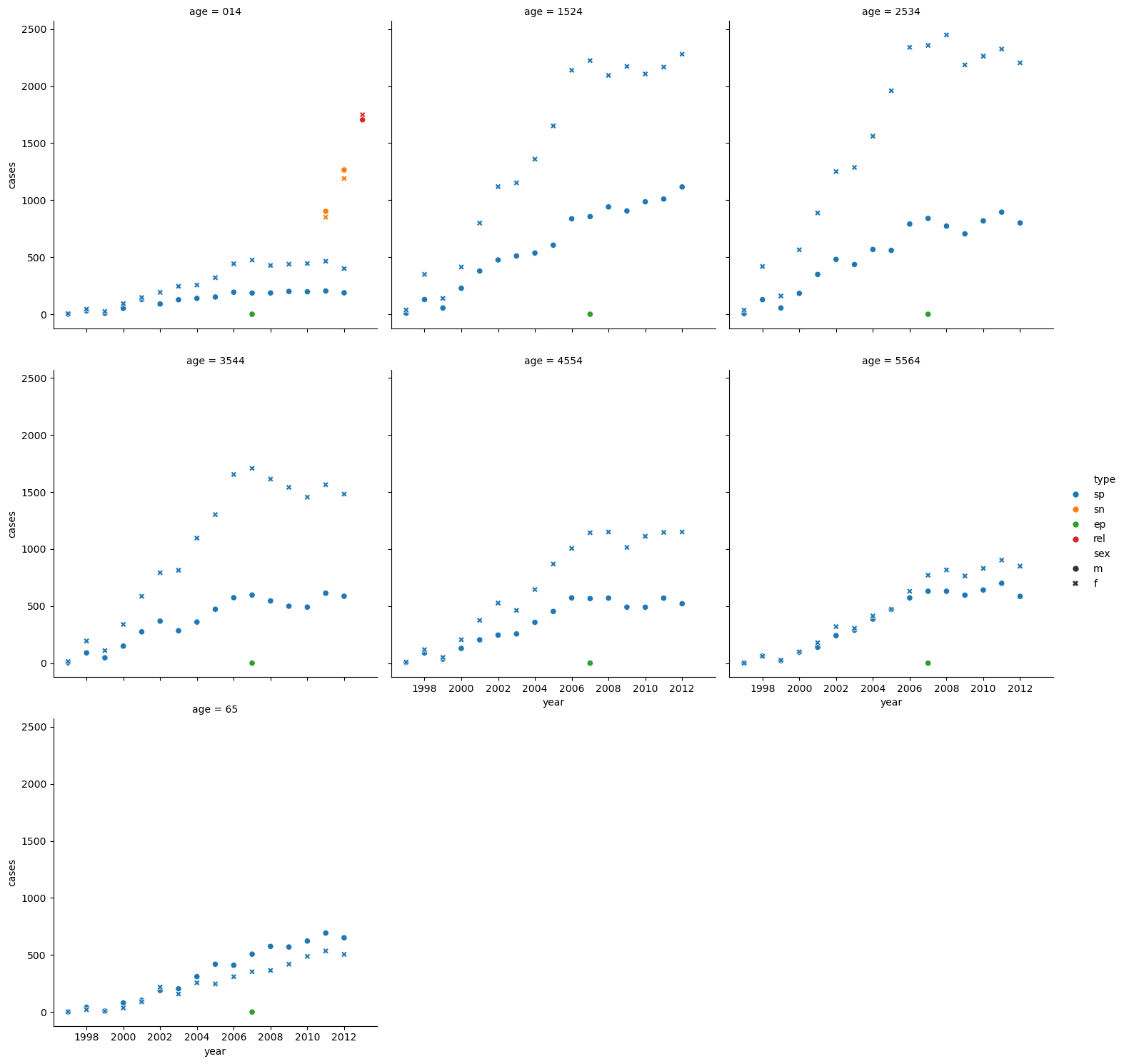
Internet data types
JSON data
JSON (short for JavaScript Object Notation) is a standard format for sending data by HTTP request between web browsers and other applications.
Here is an example from NYC Open Data
The data is on Citi-Bike Stations in NYC, and the start of it looks like:
{"data": {"stations": [{"is_installed": 0, "is_returning": 0, "num_bikes_disabled": 0, "is_renting": 0, "station_id": "1817822909864556798", "num_docks_disabled": 0, "legacy_id": "1817822909864556798", "last_reported": 1739366429, "num_ebikes_available": 0, "num_bikes_available": 0, "num_docks_available": 0, "eightd_has_available_keys": false}, {"is_installed": 1, "is_returning": 1, "num_bikes_disabled": 4, "is_renting": 1, "station_id": "5a24acbb-94ef-4cb5-9db8-f6073ac26a2b", "num_docks_disabled": 0, "legacy_id": "4302", "num_scooters_available": 0, "last_reported": 1739394755, "num_ebikes_available": 0, We can see it looks nearly like valid Python code (e.g. a dictionary).
A helpful package for reading JSON data is json:
import jsonbike_data = json.load(open('../data/citibike.json'))bike_data.keys()dict_keys(['data', 'last_updated', 'ttl', 'version'])bike_data['data'].keys()dict_keys(['stations'])stations = pd.DataFrame(bike_data['data']['stations'])stations.head()| is_installed | is_returning | num_bikes_disabled | is_renting | station_id | num_docks_disabled | legacy_id | last_reported | num_ebikes_available | num_bikes_available | num_docks_available | eightd_has_available_keys | num_scooters_available | num_scooters_unavailable | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 0 | 0 | 1817822909864556798 | 0 | 1817822909864556798 | 1739366429 | 0 | 0 | 0 | False | NaN | NaN |
| 1 | 1 | 1 | 1 | 1 | bacfe3c2-7946-45ed-91f9-f5b91791de75 | 0 | 3503 | 1739395003 | 1 | 2 | 27 | False | 0.0 | 0.0 |
| 2 | 1 | 1 | 5 | 1 | 9394fb49-583f-4426-8d2f-a2b0ffb584d2 | 0 | 3737 | 1739395004 | 4 | 34 | 10 | False | 0.0 | 0.0 |
| 3 | 1 | 1 | 9 | 1 | 1869753654337059182 | 0 | 1869753654337059182 | 1739394999 | 1 | 10 | 2 | False | 0.0 | 0.0 |
| 4 | 1 | 1 | 6 | 1 | 1f8f6714-42a7-45d5-86cd-a093c940b2c9 | 0 | 4635 | 1739395000 | 1 | 2 | 19 | False | 0.0 | 0.0 |
HTML data
To read in HTML tables, for example, we can use the pd.read_html command.
Note: we need the following packages:
conda install lxml beautifulsoup4 html5lib
This is because pd.read_html uses lxml or Beautiful Soup under the hood to parse data from HTML.
The following is from the pandas documenation, and gets a table of failed banks from the FDIC website.
url = "https://www.fdic.gov/resources/resolutions/bank-failures/failed-bank-list"
banks = pd.read_html(url)type(banks)listNote: pd.read_html reads HTML tables into a list of DataFrame objects. This is helpful if there is more than one table on the website.
banks[0]| Bank Name | City | State | Cert | Aquiring Institution | Closing Date | Fund Sort ascending | |
|---|---|---|---|---|---|---|---|
| 0 | Pulaski Savings Bank | Chicago | Illinois | 28611 | Millennium Bank | January 17, 2025 | 10548 |
| 1 | The First National Bank of Lindsay | Lindsay | Oklahoma | 4134 | First Bank & Trust Co., Duncan, OK | October 18, 2024 | 10547 |
| 2 | Republic First Bank dba Republic Bank | Philadelphia | Pennsylvania | 27332 | Fulton Bank, National Association | April 26, 2024 | 10546 |
| 3 | Citizens Bank | Sac City | Iowa | 8758 | Iowa Trust & Savings Bank | November 3, 2023 | 10545 |
| 4 | Heartland Tri-State Bank | Elkhart | Kansas | 25851 | Dream First Bank, N.A. | July 28, 2023 | 10544 |
| 5 | First Republic Bank | San Francisco | California | 59017 | JPMorgan Chase Bank, N.A. | May 1, 2023 | 10543 |
| 6 | Signature Bank | New York | New York | 57053 | Flagstar Bank, N.A. | March 12, 2023 | 10540 |
| 7 | Silicon Valley Bank | Santa Clara | California | 24735 | First Citizens Bank & Trust Company | March 10, 2023 | 10539 |
| 8 | Almena State Bank | Almena | Kansas | 15426 | Equity Bank | October 23, 2020 | 10538 |
| 9 | First City Bank of Florida | Fort Walton Beach | Florida | 16748 | United Fidelity Bank, fsb | October 16, 2020 | 10537 |
XML data
XML is another common structured data format supporting hierarchical, nested data with metadata. XML and HTML are similar, but XML is more general.
The following is an example from the xml.etree.ElementTree documentation.
!cat ../data/country_data.xml<?xml version="1.0"?>
<data>
<country name="Liechtenstein">
<rank>1</rank>
<year>2008</year>
<gdppc>141100</gdppc>
<neighbor name="Austria" direction="E"/>
<neighbor name="Switzerland" direction="W"/>
</country>
<country name="Singapore">
<rank>4</rank>
<year>2011</year>
<gdppc>59900</gdppc>
<neighbor name="Malaysia" direction="N"/>
</country>
<country name="Panama">
<rank>68</rank>
<year>2011</year>
<gdppc>13600</gdppc>
<neighbor name="Costa Rica" direction="W"/>
<neighbor name="Colombia" direction="E"/>
</country>
</data>import xml.etree.ElementTree as ETtree = ET.parse('../data/country_data.xml')
root = tree.getroot()XML documents are represented as a tree of elements
Each element has:
- a tag (name)
- attributes (optional)
- text content (optional)
root.tag'data'root.attrib{}for child in root:
print(child.tag, child.attrib, child.text)
for child2 in child:
print('--', child2.tag, child2.attrib, child2.text)
print('\n')country {'name': 'Liechtenstein'}
-- rank {} 1
-- year {} 2008
-- gdppc {} 141100
-- neighbor {'name': 'Austria', 'direction': 'E'} None
-- neighbor {'name': 'Switzerland', 'direction': 'W'} None
country {'name': 'Singapore'}
-- rank {} 4
-- year {} 2011
-- gdppc {} 59900
-- neighbor {'name': 'Malaysia', 'direction': 'N'} None
country {'name': 'Panama'}
-- rank {} 68
-- year {} 2011
-- gdppc {} 13600
-- neighbor {'name': 'Costa Rica', 'direction': 'W'} None
-- neighbor {'name': 'Colombia', 'direction': 'E'} None
We can access elements using indexing:
root[0]<Element 'country' at 0x3064bb830>root[0].attrib{'name': 'Liechtenstein'}root[0].attrib['name']'Liechtenstein'# We can also use .get to extract a specific attribute value
root[0].get('name') 'Liechtenstein'# We can use .find to get the first matching subelement
root[0].find('rank')<Element 'rank' at 0x3064b9990># .find only works one subelement down - use the path for deeper elements
root.find('country/rank')<Element 'rank' at 0x3064b9990># if you don't know the path, use './/tag_name'
root.find('.//rank')<Element 'rank' at 0x3064b9990># We can use .find to get a list of all matching subelements
root.findall('.//rank')[<Element 'rank' at 0x3064b9990>,
<Element 'rank' at 0x3064b84f0>,
<Element 'rank' at 0x306499cb0>]You can also use pd.read_xml, however, this method is best designed to import shallow XML documents.
This function will always return a single DataFrame or raise exceptions due to issues with XML document, xpath, or other parameters.
xml_df = pd.read_xml('../data/country_data.xml')xml_df| name | rank | year | gdppc | neighbor | |
|---|---|---|---|---|---|
| 0 | Liechtenstein | 1 | 2008 | 141100 | NaN |
| 1 | Singapore | 4 | 2011 | 59900 | NaN |
| 2 | Panama | 68 | 2011 | 13600 | NaN |
Note that we lose the neighbor information.
More specifically, these are the kinds of formats easiest to read with pd.read_xml
<root>
<row>
<column1>data</column1>
<column2>data</column2>
<column3>data</column3>
...
</row>
<row>
...
</row>
...
</root>Illustration: Exchange Rate Data
Let’s look again at the exchange rate data from Lectures 2 and 3, but this time in XML form.
df = pd.read_xml('../data/eurofxref-hist-90d.xml')df| subject | name | Cube | |
|---|---|---|---|
| 0 | Reference rates | None | None |
| 1 | None | European Central Bank | None |
| 2 | None | None | \n |
The structure is too complicated to be directly read with pd.read_xml.
tree = ET.parse('../data/eurofxref-hist-90d.xml')
root = tree.getroot()
root.tag'{http://www.gesmes.org/xml/2002-08-01}Envelope'for child in root:
print(child.tag, child.attrib, child.text){http://www.gesmes.org/xml/2002-08-01}subject {} Reference rates
{http://www.gesmes.org/xml/2002-08-01}Sender {}
{http://www.ecb.int/vocabulary/2002-08-01/eurofxref}Cube {}
for child in root:
print(child.tag, child.attrib, child.text)
for i, child2 in enumerate(child):
if i < 5:
print('---', child2.tag, child2.attrib, child2.text){http://www.gesmes.org/xml/2002-08-01}subject {} Reference rates
{http://www.gesmes.org/xml/2002-08-01}Sender {}
--- {http://www.gesmes.org/xml/2002-08-01}name {} European Central Bank
{http://www.ecb.int/vocabulary/2002-08-01/eurofxref}Cube {}
--- {http://www.ecb.int/vocabulary/2002-08-01/eurofxref}Cube {'time': '2024-04-16'}
--- {http://www.ecb.int/vocabulary/2002-08-01/eurofxref}Cube {'time': '2024-04-15'}
--- {http://www.ecb.int/vocabulary/2002-08-01/eurofxref}Cube {'time': '2024-04-12'}
--- {http://www.ecb.int/vocabulary/2002-08-01/eurofxref}Cube {'time': '2024-04-11'}
--- {http://www.ecb.int/vocabulary/2002-08-01/eurofxref}Cube {'time': '2024-04-10'}
root[2].tag '{http://www.ecb.int/vocabulary/2002-08-01/eurofxref}Cube'cube = root[2]for i, child in enumerate(cube):
if i < 5:
print(child.attrib)
for j, child2 in enumerate(child):
if j < 5:
print('----', child2.attrib){'time': '2024-04-16'}
---- {'currency': 'USD', 'rate': '1.0637'}
---- {'currency': 'JPY', 'rate': '164.54'}
---- {'currency': 'BGN', 'rate': '1.9558'}
---- {'currency': 'CZK', 'rate': '25.21'}
---- {'currency': 'DKK', 'rate': '7.4609'}
{'time': '2024-04-15'}
---- {'currency': 'USD', 'rate': '1.0656'}
---- {'currency': 'JPY', 'rate': '164.05'}
---- {'currency': 'BGN', 'rate': '1.9558'}
---- {'currency': 'CZK', 'rate': '25.324'}
---- {'currency': 'DKK', 'rate': '7.4606'}
{'time': '2024-04-12'}
---- {'currency': 'USD', 'rate': '1.0652'}
---- {'currency': 'JPY', 'rate': '163.16'}
---- {'currency': 'BGN', 'rate': '1.9558'}
---- {'currency': 'CZK', 'rate': '25.337'}
---- {'currency': 'DKK', 'rate': '7.4603'}
{'time': '2024-04-11'}
---- {'currency': 'USD', 'rate': '1.0729'}
---- {'currency': 'JPY', 'rate': '164.18'}
---- {'currency': 'BGN', 'rate': '1.9558'}
---- {'currency': 'CZK', 'rate': '25.392'}
---- {'currency': 'DKK', 'rate': '7.4604'}
{'time': '2024-04-10'}
---- {'currency': 'USD', 'rate': '1.086'}
---- {'currency': 'JPY', 'rate': '164.89'}
---- {'currency': 'BGN', 'rate': '1.9558'}
---- {'currency': 'CZK', 'rate': '25.368'}
---- {'currency': 'DKK', 'rate': '7.4594'}cube[0][0].attrib['currency']'USD'cube[0][0].attrib['rate']'1.0637'xml_dict = {}
for i, child in enumerate(cube):
temp_dict = {}
for j, child2 in enumerate(child):
temp_dict[child2.attrib['currency']] = child2.attrib['rate']
xml_dict[child.attrib['time']] = temp_dictdf_xml = pd.DataFrame(xml_dict)df_xml.head()| 2024-04-16 | 2024-04-15 | 2024-04-12 | 2024-04-11 | 2024-04-10 | 2024-04-09 | 2024-04-08 | 2024-04-05 | 2024-04-04 | 2024-04-03 | ... | 2024-01-31 | 2024-01-30 | 2024-01-29 | 2024-01-26 | 2024-01-25 | 2024-01-24 | 2024-01-23 | 2024-01-22 | 2024-01-19 | 2024-01-18 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| USD | 1.0637 | 1.0656 | 1.0652 | 1.0729 | 1.086 | 1.0867 | 1.0823 | 1.0841 | 1.0852 | 1.0783 | ... | 1.0837 | 1.0846 | 1.0823 | 1.0871 | 1.0893 | 1.0905 | 1.0872 | 1.089 | 1.0887 | 1.0875 |
| JPY | 164.54 | 164.05 | 163.16 | 164.18 | 164.89 | 164.97 | 164.43 | 164.1 | 164.69 | 163.66 | ... | 160.19 | 159.97 | 160.13 | 160.62 | 160.81 | 160.46 | 160.88 | 160.95 | 161.17 | 160.89 |
| BGN | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | ... | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 | 1.9558 |
| CZK | 25.21 | 25.324 | 25.337 | 25.392 | 25.368 | 25.38 | 25.354 | 25.286 | 25.322 | 25.352 | ... | 24.891 | 24.831 | 24.806 | 24.748 | 24.756 | 24.786 | 24.824 | 24.758 | 24.813 | 24.734 |
| DKK | 7.4609 | 7.4606 | 7.4603 | 7.4604 | 7.4594 | 7.459 | 7.4588 | 7.459 | 7.4589 | 7.4589 | ... | 7.455 | 7.4543 | 7.4538 | 7.4549 | 7.456 | 7.4568 | 7.4574 | 7.4585 | 7.4575 | 7.4571 |
5 rows × 62 columns
df_xml = df_xml.Tdf_xml.head()| USD | JPY | BGN | CZK | DKK | GBP | HUF | PLN | RON | SEK | ... | ILS | INR | KRW | MXN | MYR | NZD | PHP | SGD | THB | ZAR | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2024-04-16 | 1.0637 | 164.54 | 1.9558 | 25.21 | 7.4609 | 0.8544 | 394.63 | 4.3435 | 4.9764 | 11.635 | ... | 4.0001 | 88.8975 | 1481.62 | 17.9141 | 5.0973 | 1.8072 | 60.561 | 1.4509 | 38.958 | 20.2104 |
| 2024-04-15 | 1.0656 | 164.05 | 1.9558 | 25.324 | 7.4606 | 0.85405 | 394.25 | 4.2938 | 4.9742 | 11.5583 | ... | 3.9578 | 88.895 | 1473.79 | 17.6561 | 5.0925 | 1.7943 | 60.558 | 1.4498 | 39.124 | 20.2067 |
| 2024-04-12 | 1.0652 | 163.16 | 1.9558 | 25.337 | 7.4603 | 0.85424 | 391.63 | 4.2603 | 4.9717 | 11.5699 | ... | 4.0175 | 88.914 | 1469.79 | 17.5982 | 5.081 | 1.7872 | 60.256 | 1.4478 | 38.853 | 19.9907 |
| 2024-04-11 | 1.0729 | 164.18 | 1.9558 | 25.392 | 7.4604 | 0.85525 | 389.8 | 4.257 | 4.9713 | 11.531 | ... | 4.03 | 89.4385 | 1469.32 | 17.6448 | 5.0936 | 1.792 | 60.577 | 1.4518 | 39.22 | 20.1614 |
| 2024-04-10 | 1.086 | 164.89 | 1.9558 | 25.368 | 7.4594 | 0.85515 | 389.18 | 4.2563 | 4.969 | 11.4345 | ... | 4.0324 | 90.3585 | 1467.33 | 17.7305 | 5.1558 | 1.7856 | 61.408 | 1.4605 | 39.536 | 20.0851 |
5 rows × 30 columns
df_xml.index.name = 'Time'df_xml = df_xml.reset_index()df_xml.head()| Time | USD | JPY | BGN | CZK | DKK | GBP | HUF | PLN | RON | ... | ILS | INR | KRW | MXN | MYR | NZD | PHP | SGD | THB | ZAR | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 2024-04-16 | 1.0637 | 164.54 | 1.9558 | 25.21 | 7.4609 | 0.8544 | 394.63 | 4.3435 | 4.9764 | ... | 4.0001 | 88.8975 | 1481.62 | 17.9141 | 5.0973 | 1.8072 | 60.561 | 1.4509 | 38.958 | 20.2104 |
| 1 | 2024-04-15 | 1.0656 | 164.05 | 1.9558 | 25.324 | 7.4606 | 0.85405 | 394.25 | 4.2938 | 4.9742 | ... | 3.9578 | 88.895 | 1473.79 | 17.6561 | 5.0925 | 1.7943 | 60.558 | 1.4498 | 39.124 | 20.2067 |
| 2 | 2024-04-12 | 1.0652 | 163.16 | 1.9558 | 25.337 | 7.4603 | 0.85424 | 391.63 | 4.2603 | 4.9717 | ... | 4.0175 | 88.914 | 1469.79 | 17.5982 | 5.081 | 1.7872 | 60.256 | 1.4478 | 38.853 | 19.9907 |
| 3 | 2024-04-11 | 1.0729 | 164.18 | 1.9558 | 25.392 | 7.4604 | 0.85525 | 389.8 | 4.257 | 4.9713 | ... | 4.03 | 89.4385 | 1469.32 | 17.6448 | 5.0936 | 1.792 | 60.577 | 1.4518 | 39.22 | 20.1614 |
| 4 | 2024-04-10 | 1.086 | 164.89 | 1.9558 | 25.368 | 7.4594 | 0.85515 | 389.18 | 4.2563 | 4.969 | ... | 4.0324 | 90.3585 | 1467.33 | 17.7305 | 5.1558 | 1.7856 | 61.408 | 1.4605 | 39.536 | 20.0851 |
5 rows × 31 columns
df_xml['Time'] = pd.to_datetime(df_xml['Time'])df_xml = df_xml.melt(id_vars=['Time'],
var_name = 'currency',
value_name='rate')df_xml| Time | currency | rate | |
|---|---|---|---|
| 0 | 2024-04-16 | USD | 1.0637 |
| 1 | 2024-04-15 | USD | 1.0656 |
| 2 | 2024-04-12 | USD | 1.0652 |
| 3 | 2024-04-11 | USD | 1.0729 |
| 4 | 2024-04-10 | USD | 1.086 |
| ... | ... | ... | ... |
| 1855 | 2024-01-24 | ZAR | 20.5366 |
| 1856 | 2024-01-23 | ZAR | 20.7022 |
| 1857 | 2024-01-22 | ZAR | 20.8846 |
| 1858 | 2024-01-19 | ZAR | 20.6892 |
| 1859 | 2024-01-18 | ZAR | 20.597 |
1860 rows × 3 columns