import torch
import torch.nn as nn
import torch.optim as optim
import numpy as np
import matplotlib.pyplot as plt
import scipy
from scipy.stats import t
from scipy.stats import ttest_1samp
torch.manual_seed(42)<torch._C.Generator at 0x12031c3f0>import torch
import torch.nn as nn
import torch.optim as optim
import numpy as np
import matplotlib.pyplot as plt
import scipy
from scipy.stats import t
from scipy.stats import ttest_1samp
torch.manual_seed(42)<torch._C.Generator at 0x12031c3f0>Let’s toss a coin 10 times.
Let’s repeat this 10,000 times.
How many times will we see 10 heads and 0 tails?
If we do enough tests, we will see rare things, even when the null hypothesis is true!
How do we account for this?
We run one-sample t-tests for m hypotheses.
We do the following corrections:
array([ 1.1590115 , 0.29437799, -1.20388884, -1.39994129, 0.80287114,
-0.66532837, 0.94760322, -1.02684096, -0.53667259, 1.26035408,
0.30475883, -0.68428187, -0.7424497 , -1.19441991, -0.6307807 ,
1.08867092, 0.65729084, 0.02539257, 1.05287459, -1.93841006,
2.17832052, -2.4701497 , 2.02294352, 0.06927303, 0.54318123,
1.9602065 , -0.35964106, 1.04538793, -1.52884496, 0.34965436,
-1.33769048, 0.6165512 , 0.67023305, -1.11043292, 0.9638287 ,
-0.46430343, -0.41250436, -0.30909244, -0.02039219, -0.3204189 ])def ttest_1(x, h0_mean=0):
df = n-1
mean = x.mean() # sample mean x_bar
d = mean - h0_mean # x_bar - mu (mu=0 under H_0)
v = np.var(x) # sample variance
denom = np.sqrt(v / n) # variance of sample mean
tstat = np.abs(d / denom)
# xmean - h0_mean / (sqrt(var/n))
# our test-stat is a t distributed random variable with n-1 degrees of freedom
pval = t.cdf(-tstat, df = df) + (1 - t.cdf(tstat, df = df))
# pval - probability in the lower and upper tails of our t distribution
return pval# no multiple testing correction
# we expect to reject m * 0.05 samples
alpha = 0.05
nmp = np.where(pvals < alpha)[0]
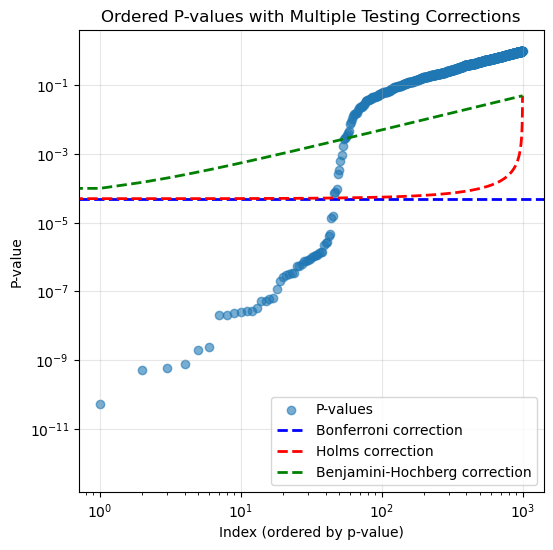
print("No multiple testing correction: reject ", nmp.shape[0])No multiple testing correction: reject 45Bonferroni: reject 0# holms
ord_pvals = np.argsort(pvals)
holms = []
for j, s in enumerate(ord_pvals):
#j = 0, s is index of smallest p-val
denom = m - j
if pvals[s] <= (alpha/denom):
holms.append(s)
else:
break
print("Holms: reject ", len(holms))Holms: reject 0# BH procedure # this is different from holms and bonferroni in that
# we control FDR, not FWER
q = 0.05
bh = []
for j, s in enumerate(ord_pvals):
val = q * (j + 1) /m # zero indexing
if pvals[s] <= val:
bh.append(s)
else:
break
print("Benjamini-Hochberg: reject ", len(bh))Benjamini-Hochberg: reject 0plt.figure(figsize=(6, 6))
plt.scatter(range(m), pvals[ord_pvals], alpha=0.6, label='P-values')
plt.axhline(y=alpha/m, color='b', linestyle='--', label='Bonferroni correction', linewidth=2)
# Holms correction line
holms_threshold = alpha / (m - np.arange(m))
plt.plot(range(m), holms_threshold, 'r--', label='Holms correction', linewidth=2)
# Benjamini-Hochberg correction line
bh_threshold = (alpha / m) * (np.arange(m) + 1)
plt.plot(range(m), bh_threshold, 'g--', label='Benjamini-Hochberg correction', linewidth=2)
plt.xlabel('Index (ordered by p-value)')
plt.ylabel('P-value')
plt.legend()
plt.yscale('log')
plt.xscale('log')
plt.title('Ordered P-values with Multiple Testing Corrections')
plt.grid(True, alpha=0.3)
plt.show()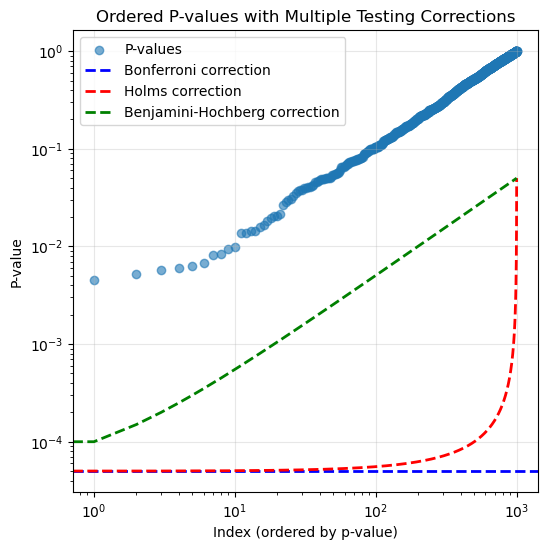
true_mean = np.array([1.0] * int(m*5/100) + [0] * int(m * 95/100))
X = np.random.normal(size = (n, m))
X = X + true_mean
plt.scatter(range(m), X.mean(axis=0))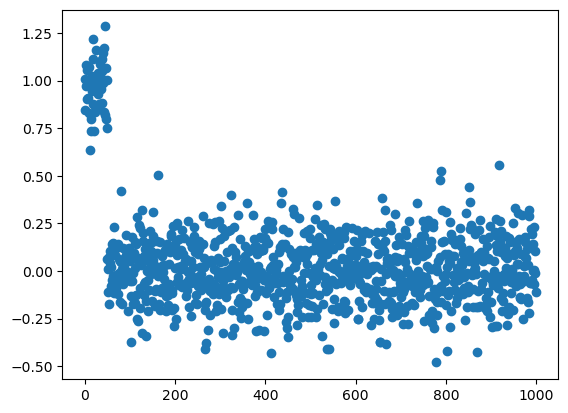
Number of hypotheses we should reject: 50# no multiple testing correction
alpha = 0.05
nmp = np.where(pvals < alpha)[0]
print("No multiple testing correction: reject ", nmp.shape[0])No multiple testing correction: reject 94Bonferroni: reject 46
# holms
ord_pvals = np.argsort(pvals)
holms = []
for j, s in enumerate(ord_pvals):
denom = m - j
if pvals[s] <= (alpha/denom):
holms.append(s)
else:
break
print("Holms: reject ", len(holms))Holms: reject 46# BH procedure
q = 0.05
bh = []
for j, s in enumerate(ord_pvals):
val = q * (j + 1) /m # zero indexing
if pvals[s] <= val:
bh.append(s)
else:
break
print("Benjamini-Hochberg: reject ", len(bh))Benjamini-Hochberg: reject 55plt.figure(figsize=(6, 6))
plt.scatter(range(m), pvals[ord_pvals], alpha=0.6, label='P-values')
plt.axhline(y=alpha/m, color='b', linestyle='--', label='Bonferroni correction', linewidth=2)
# Holms correction line
holms_threshold = alpha / (m - np.arange(m))
plt.plot(range(m), holms_threshold, 'r--', label='Holms correction', linewidth=2)
# Benjamini-Hochberg correction line
bh_threshold = (alpha / m) * (np.arange(m) + 1)
plt.plot(range(m), bh_threshold, 'g--', label='Benjamini-Hochberg correction', linewidth=2)
plt.xlabel('Index (ordered by p-value)')
plt.ylabel('P-value')
plt.yscale('log')
plt.xscale('log')
plt.legend()
plt.title('Ordered P-values with Multiple Testing Corrections')
plt.grid(True, alpha=0.3)
plt.show()